Technologies for massive parallel sequencing
🧬
Daniel Fürth
Assistant professor
SciLifeLab/Uppsala University
🔗 furthlab.xyz
@furthlab
Technologies for massive parallel sequencing
Daniel Fürth
Assistant professor
SciLifeLab/Uppsala University

🔗 furthlab.xyz
@furthlab
Monday 23 Jan. A7:103, BMC, 15:15 - 17:00
📚🙇🏽♀️🙇🏻♂️📖 Lärandemål
Kurplan: 🔗3MG010
Innehåll:
- Kromosomstruktur, normal och avvikande kromosomuppsättning.
- Monogen och multifaktoriell nedärvning.
- Metoder för gen-identifiering vid enkla och komplexa egenskaper.
- Populationsgenetik, riskberäkning i familjer.
- Diagnostik inom klinisk genetik, screening av nyfödda samt bärare.
- Det humana genomets sammansättning och genetisk variation.
- Genomevolution, genetiska modellorganismer och komparativ genomik.
- 🧬
Metoder att analysera hela genoms struktur och funktion, storskalig analys av DNA-sekvens och epigenetisk variation, samt mätning av transkriptions- och proteinnivåer. - Metoder för att koppla en gen till en sjukdom.
- Mekanismer för reglering av geners uttryck.
- Användandet av genetiska markörer inom forensisk medicin. Etiska principer, processer och deklarationer.
- Farmakogenetik och cancergenetik.
📚🙇🏽♀️🙇🏻♂️📖 Lärandemål
Saker som vi ska gå igenom idag:
- 📜 Historiskt perspektiv:
- från Sanger-sekvensering till Next-Generation Sequencing (NGS)
- 🔴🟢🟣🔵 Sanger-sekvensering genom kapillärelektrofores
- NGS solid-phase substrate:
- 🪨 Emulsion PCR
- 🪨 Bridge amplification
- NGS primer extension:
- 🧪 Sequencing by Synthesis (SBS)
- 🧪 Pyrosequencing
- 🧪 Reversible-dye terminators (Illumina)
- 🧪 Sequencing by Ligation (SBL)
- 🧬 Long read sequencing
- PacBio
- NanoPore
📜 Sanger sequencing ➡️ Next-Generation Sequencing (NGS)
Separation chemistry

Electrophoretic machine as designed by Tiselius
Separation chemistry
Polyacrylamide gel electrophoresis (PAGE)
- Length
- Charge
- Confirmation
Molecules can be run in their “native state”
preserving higher-order structure.
Or a chemical denaturant can be added to
remove structure (urea for 🧬).
Native vs non-native PAGE.
Pore size controled by % of acrylamide.
📏 Resolution:
100-1000 nt down to single-nucleotide.

Fredrick Sanger
- One of the few who received two Noble Prizes.
- 💉1958
“structure of proteins, especially that of insulin”
- 🧬1980
“determination of base sequences in nucleic acids”
- 💉1958

📜 Sanger sequencing ➡️ single-cell methods
🇬🇧 Medical Research Council
Laboratory of Molecular Biology

🏅Sydney Brenner (2002).
The golden era ⚜️⚱️🏆 of molecular biology 🧬.
🛠🧰 Restriction enzymes 🧬
🇺🇸 Restriction and modification enzymes 🇨🇭
🏅Noble Prize: 🇺🇸 Hamilton O. Smith, 🇺🇸 Daniel Nathans, and 🇨🇭 Werner Arber.

Structure of a Ribonucleic Acid

Structure of a Ribonucleic Acid
RNase A (Bovine pancreas ribonuclease):
- Only active on RNA.
- Very sturdy, survives boiling etc 🔥.
- Cleaves 3’-end of unpaired C and U residues (pyrimidine).
RNase T1 (Taka-Diastase ribonuclease T1):
- Single-stranded (ssDNA or RNA) specific exonuclease
- Catalyzes the removal of nucleotides from linear single-stranded DNA or RNA in the 3’ to 5’ direction
Nuclease Protection Assay (NPA)
A proto sequencing method.

Two-dimension partition sequencing
A proto sequencing method.

Two-dimension partition sequencing

Two-dimension partition sequencing

dNTP incorporation has base specificity

The birth of the “-”-approach of Sanger.
First sequence of a single gene

- Bacteriophage MS2
- Circular ssDNA genome.
Sanger’s Plus/Minus sequencing


Dideoxynucleotides (ddNTP) are incorporated
Polymerases can incorporate ddNTPs.
Leading to a single base extension followed by termination
No 3’hydroxyl group (3’OH).
🏆 Chain-termination method 🏅
“Sanger sequencing”

Second Noble Prize
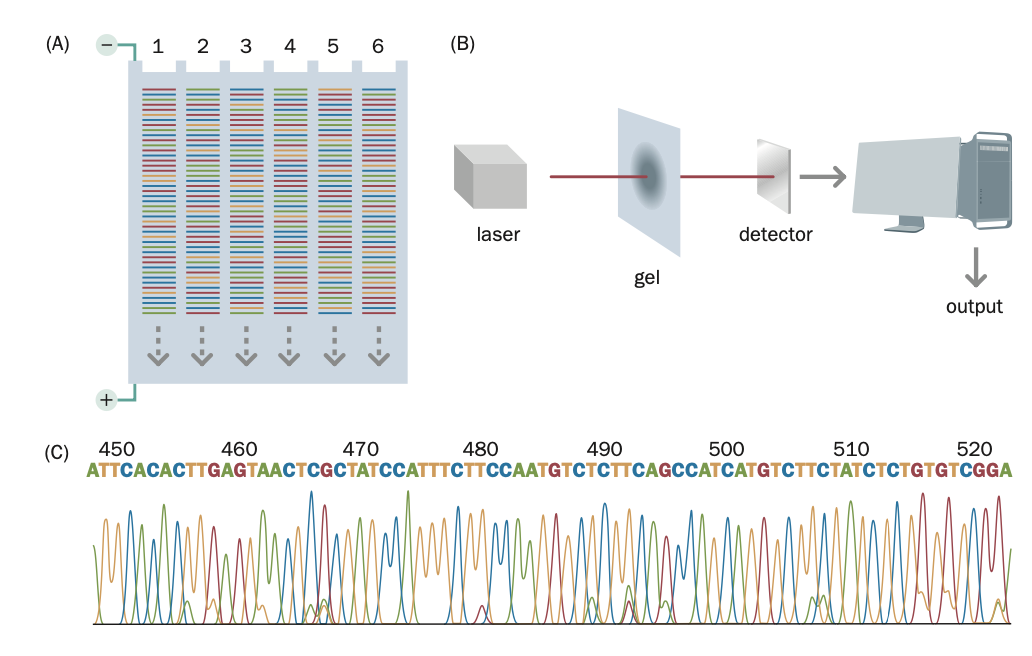
Sanger sequencing


Capillary gel electrophoresis
Maxam & Gilbert sequencing
Radioactive 5’labeling
Chemical cleavage
PAGE
The birth of biotech
California. Genentech and recombinant technologies were already creating value. Flow cytometer and Beckman Coulter commercial machines.

⛰ Cal. Tech. 👥 Leroy Hood and Mike Hunkapiller 🏢 Applied Bioscience Inc. (ABI)
Human Genome Project
Meetings that changed the world:
Santa Fe Workshop 1986.
Technological landscape
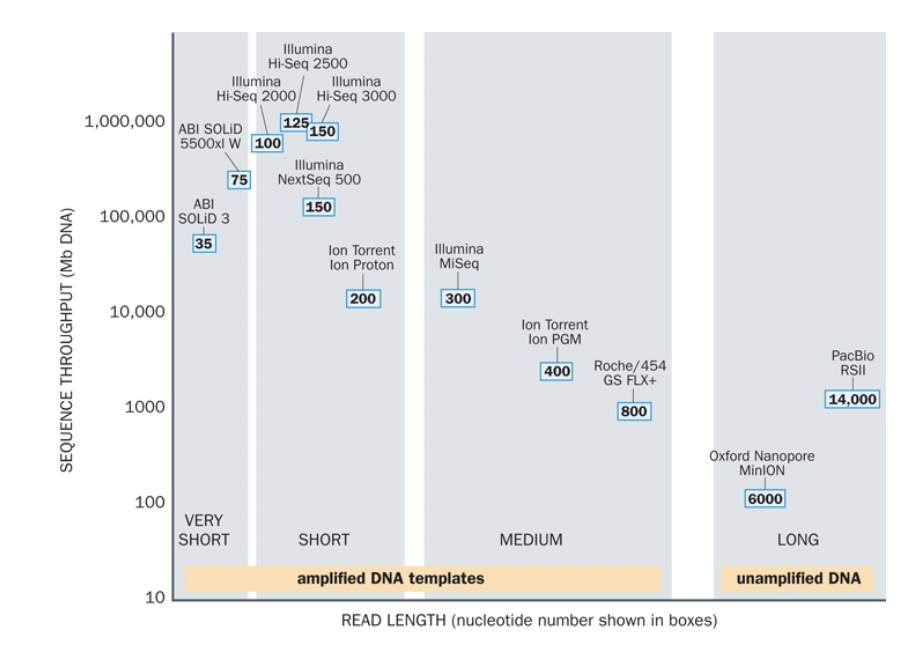
- Read length
- Throughput
Read length

Throughput
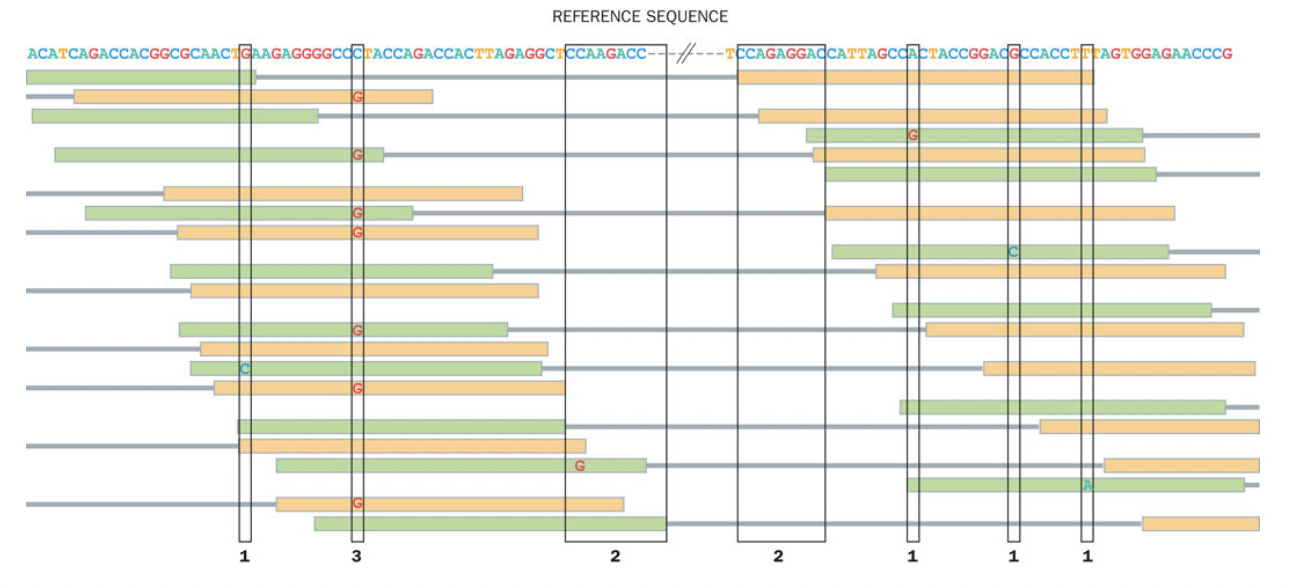
Alignment
Next-generation sequencing
All about how to massively multiplex sequencing.
Pyrosequencing
Sequencing by ligation
Reversible dye-terminators
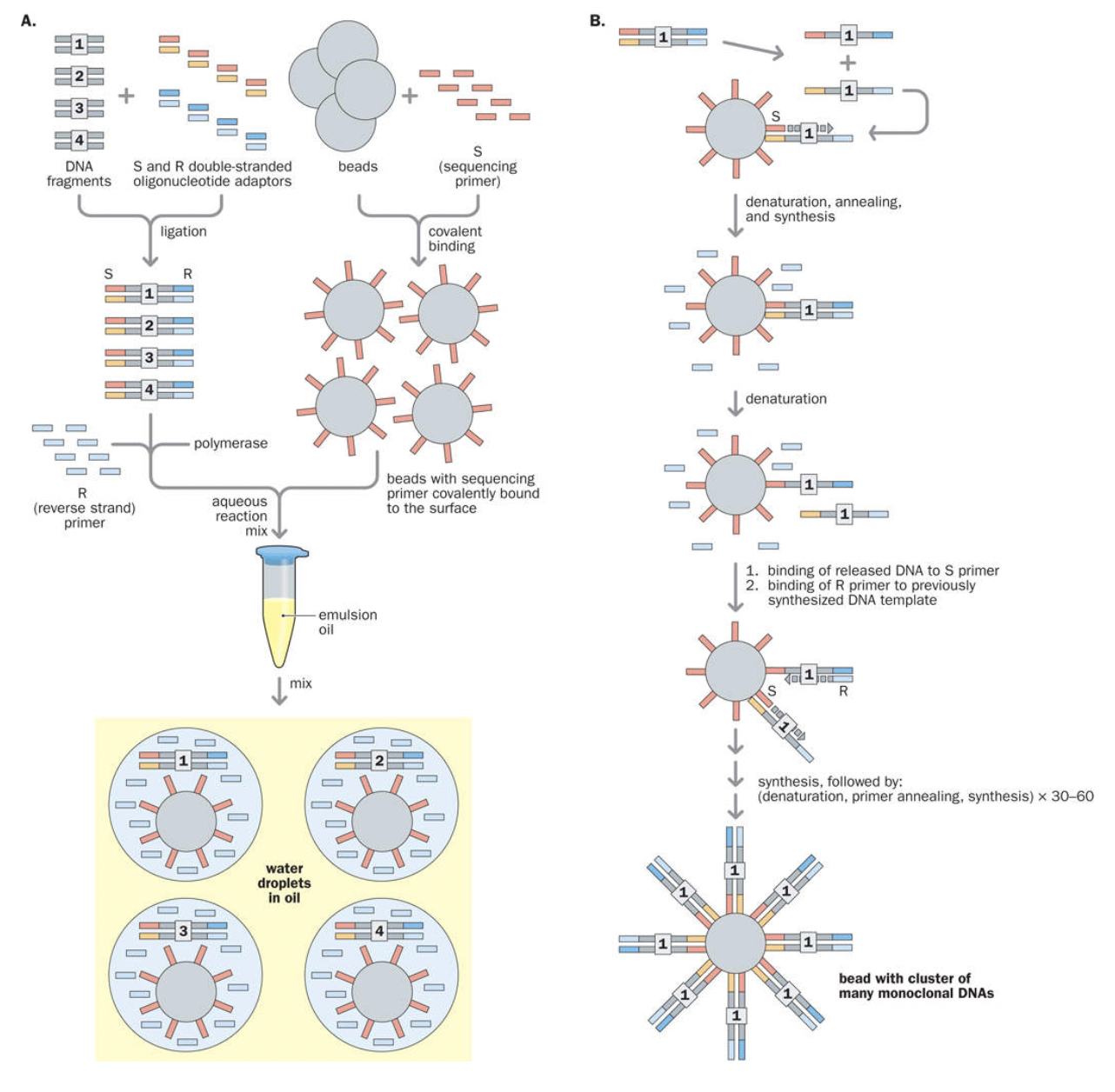
Solid-phase substrate
Bridge amplification
Emulsion PCR
DNA nanoballs
Primer extension
Sequencing by synthesis
(SBS)Sequencing by ligation
(SBL)
Pyrosequencing

Emulsion PCR/Polony sequencing

Emulsion PCR
Solexa/Illumina sequencing
- Solid support: PCR bridge amplification
- Primer extension: Reversible dye-terminators (
patent 444 about to expire!)

Solexa/Illumina sequencing
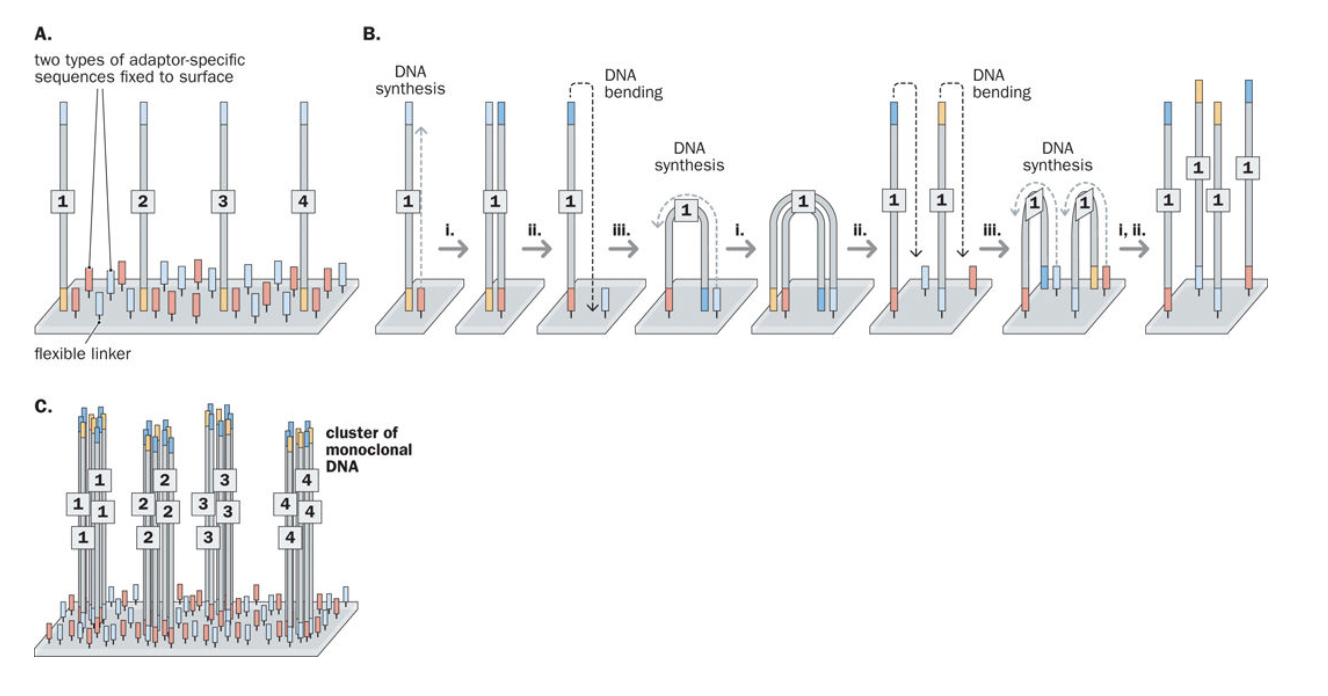
Bridge amplification
Solexa/Illumina sequencing

Solexa/Illumina sequencing

Sequencing by Ligation (SBL)
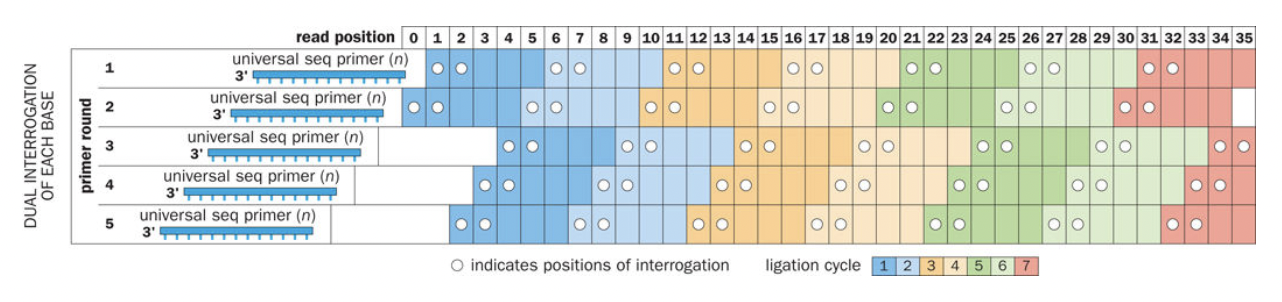
Democratization of sequencing

Paired-end and mate pair
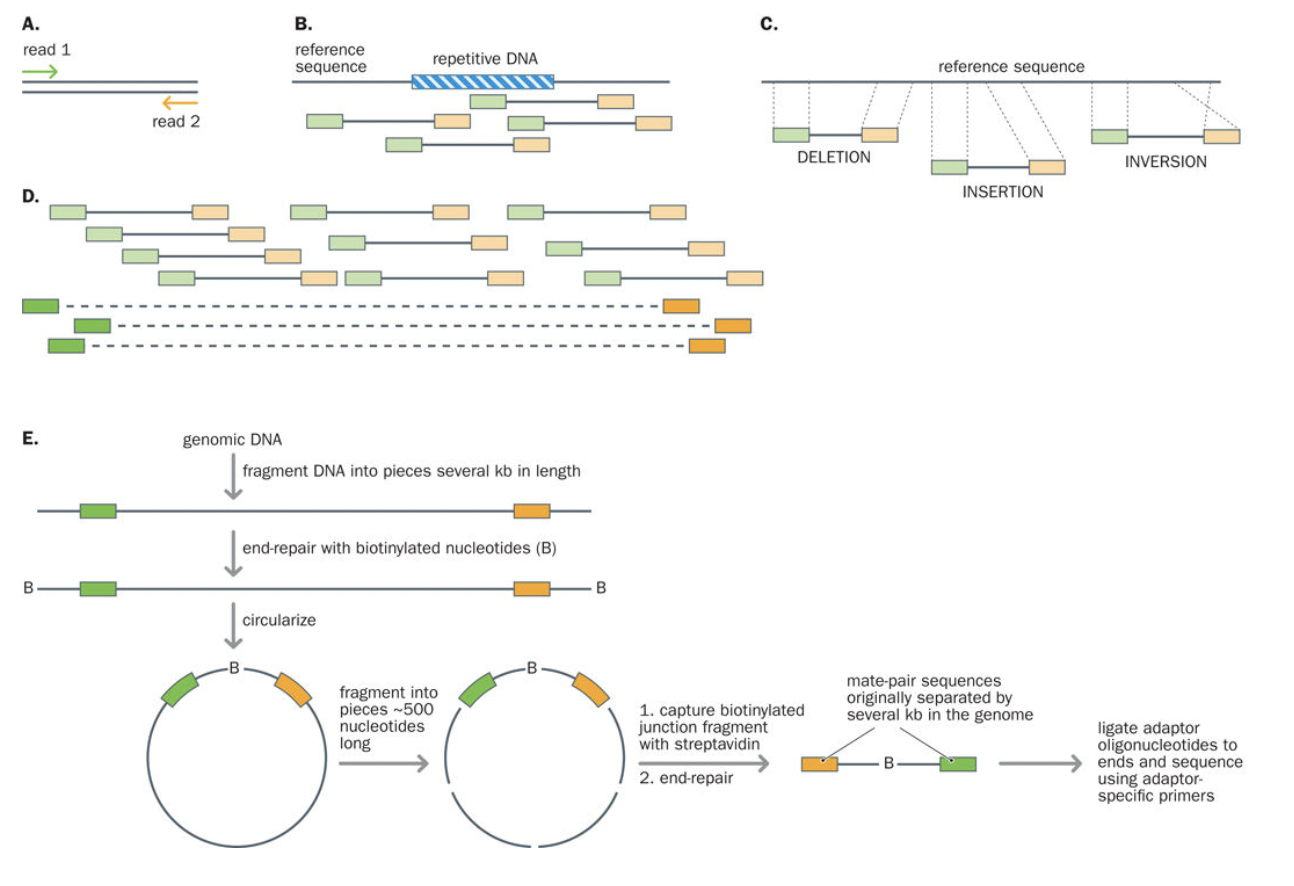
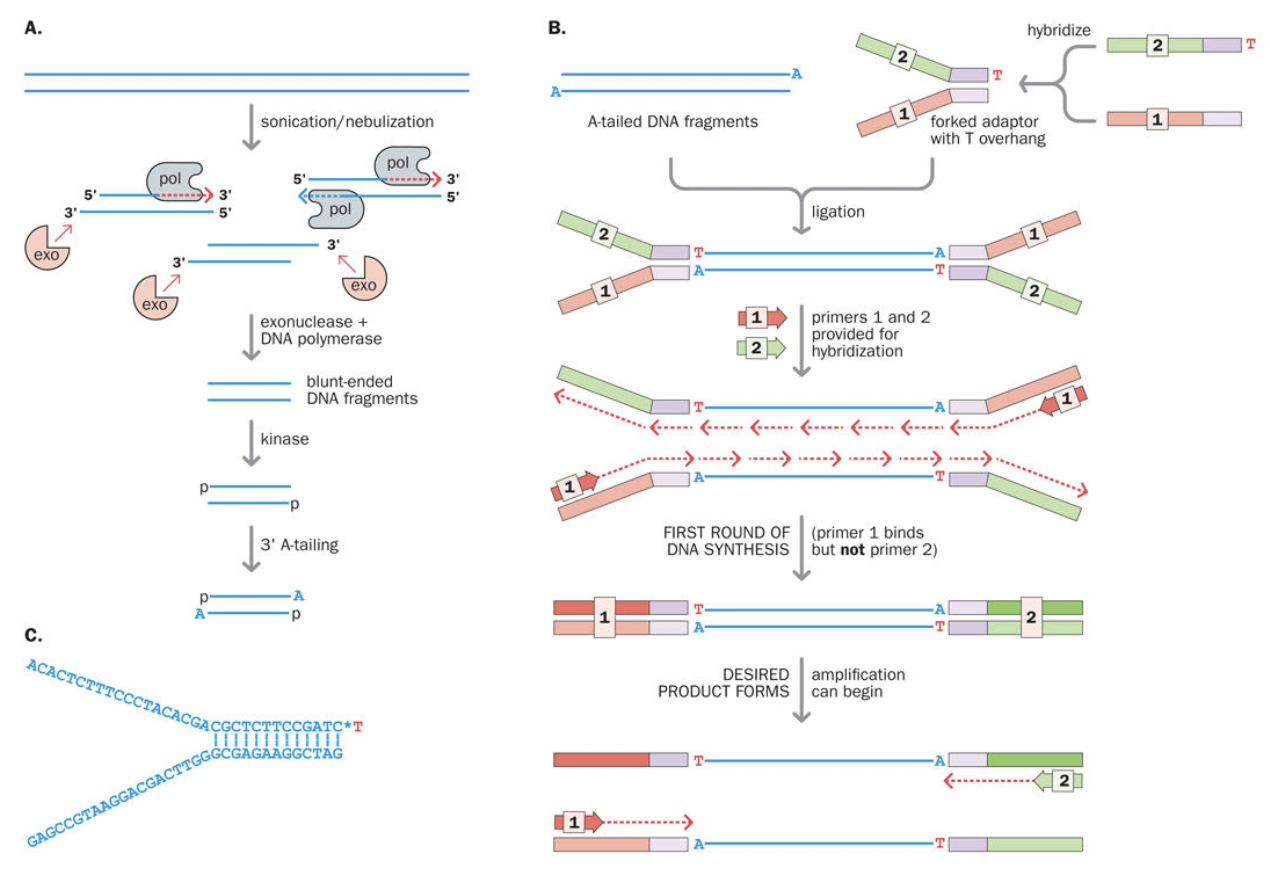
DNA library preparation
🧬 Long read sequencing
Single Molecule, Real-Time (SMRT) sequencing
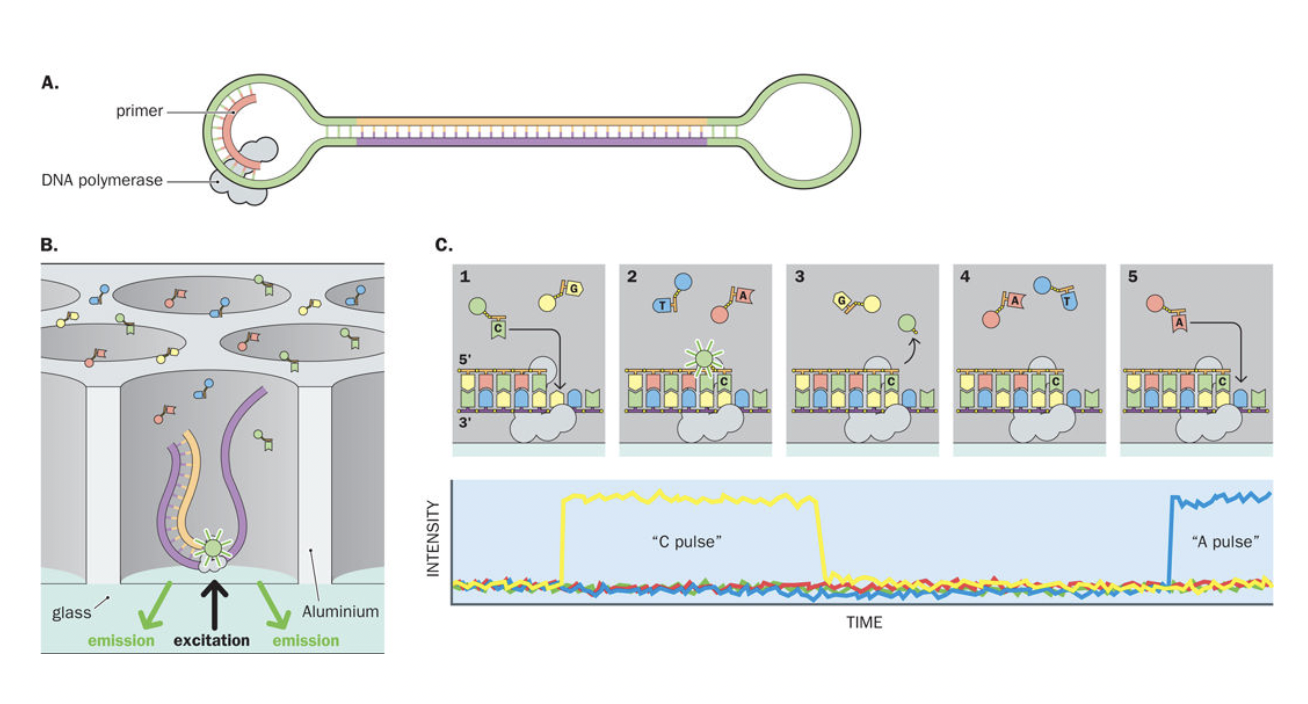
- PacBio
- SMRTbell Template Prep
- Zero-Mode Waveguides
NanoPore sequencing
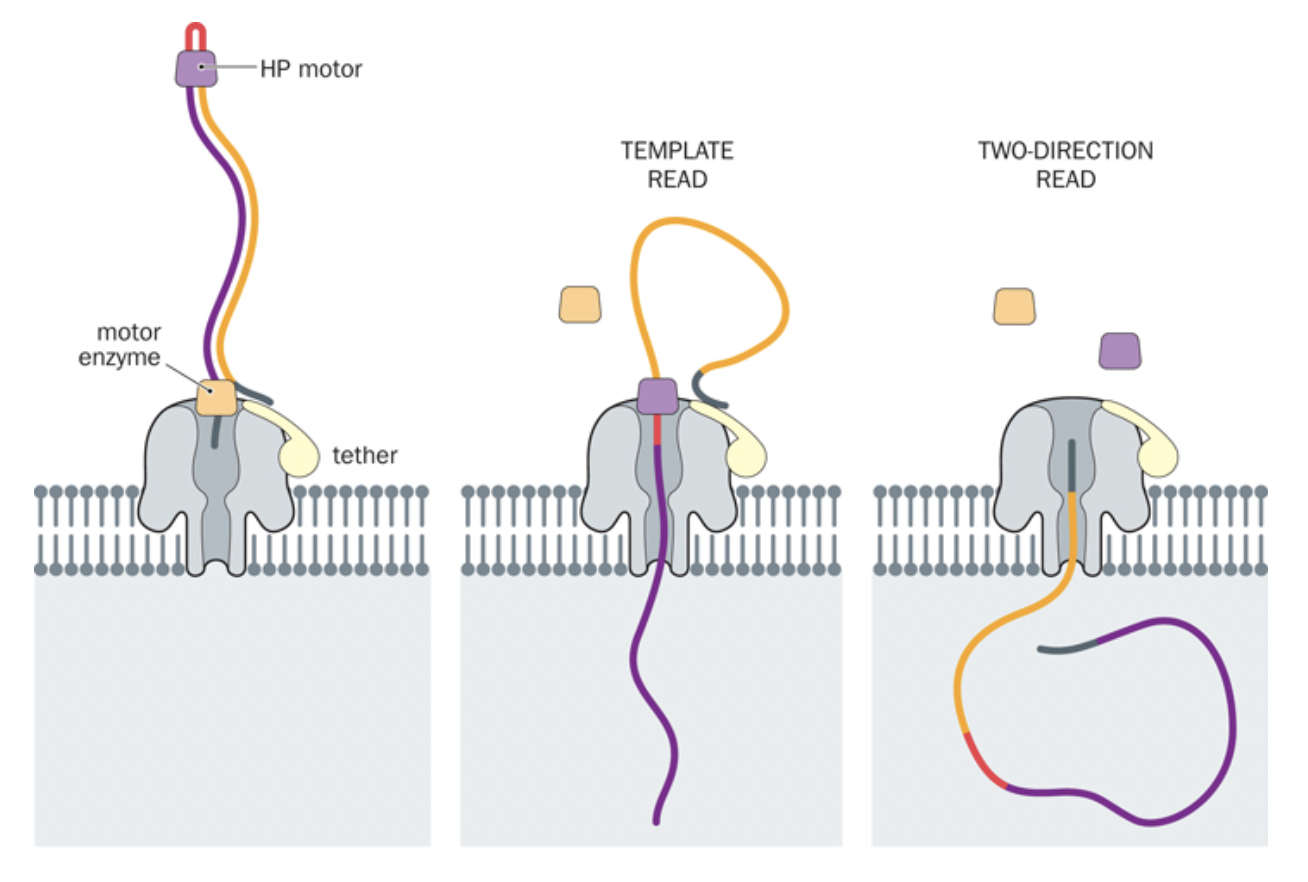
- OxfordNanopore
Unbiased in situ sequencing
In situ sequencing
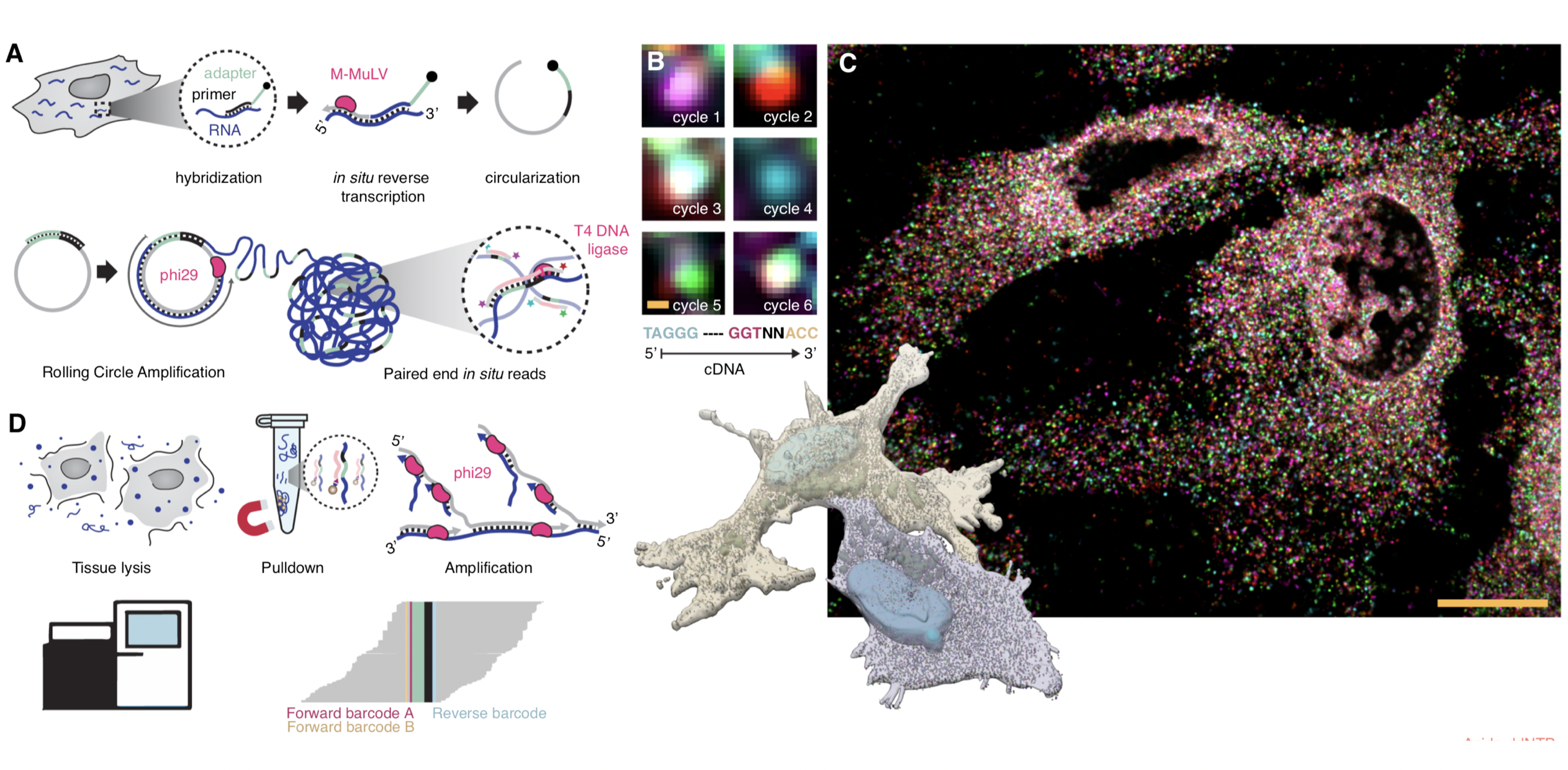
Fürth, Hatini, Lee bioRxiv

Monday 22 Jan. A7:111, BMC, 15:15 - 17:00