Our lab tries to adhere to the principle of Open Chemistry (Lavis, 2021). We aspire to share reagents and synthesize custom probes for academic scientists at cost price.
Currently, we are offering a complimentary batch of FluoroProx probes to the scientific community. First come, first served.
To request these items, simply add them to your "request cart" and proceed to checkout. Upon checkout, your selected items and shipping details will be automatically copied to your clipboard. Next, paste this information into an email and send it to us at 📨furthscilifelab.uu.se.
Please ensure to include "Sample Request" in the subject line.
By default, we ship items via regular mail, but if you have a preferred carrier, please include that information in your email.

Probes
.svg)
FluoroProx488
Amount: 10 μL @1 mM in DMSO
Reactions: 50-200 (50-200 μL Vol)
Mol. weight: 1697.82 g/mol
Ex/Em: 496 nm / 515 nm
Add to request.svg)
FluoroProx594
Amount: 10 μL @1 mM in DMSO
Reactions: 50-200 (50-200 μL Vol)
Mol. weight: 1885.13 g/mol
Ex/Em: 593 nm / 613 nm
Add to request.svg)
FluoroProx660
Amount: 10 μL @1 mM in DMSO
Reactions: 50-200 (50-200 μL Vol)
Mol. weight: 1912.25 g/mol
Ex/Em: 665 nm / 690 nm
Add to request
Molecular ruler 1.4 nm
Amount: 10 μL @1 mM in DMSO
Reactions: 50-200 (50-200 μL Vol)
Mol. weight: 675.8 g/mol
End-to-end length: ~1.4 nm
Add to request
Monosubstrate quencher
Amount: 10 μL @1 mM in DMSO
Reactions: 50-200 (50-200 μL Vol)
Mol. weight: 1027.25 g/mol
End-to-end length: ~1.4 nm
Add to requestPlasmids
This section serves as a reference for lab members to access information on plasmid availability and microscope images. It aids in anticipating experimental outcomes such as expression and localization. For plasmid distribution, please refer to Addgene.
| Plasmid | ID | Description | Image |
|---|---|---|---|
pSF-aTubCaud-ASAP5 |
TBD | ASAP5 is a non-rhodopsin-based GEVI (genetically encoded voltage indicator), making it spectrally compatible with optogenetic and rhodopsin-based tools used for perturbing activity. This specific ASAP5 is codon optimized for P.caudatum replacing rare codon usage (<20%) with frequent codons from a newly computed P.caudatum codon table (43c3d annotation/assembly v1) with 64'003 coding sequences from 18'509 transcripts compromised of 8'507'122 codons. The pAce coding sequence is inserted between two flanking 5'UTR and 3UTR from the P.caudatum alpha-tubulin gene. The vector, pSF-aTubCaud, is a modification of the original phagemid pTZ18U vector. Before transfection by microinjection the plasmid is linearized using ApaI restriction enzyme. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 30°C Strain: NEB Stable |
TBD |
pSF-aTubCaud-pAce-Kv2.1PR |
TBD | pAce-Kv2.1 PR is a plasmid designed for soma-targeted expression of a genetically encoded voltage indicator in Paramecium caudatum. It encodes pAce, a positively tuned variant of the Ace rhodopsin-based voltage sensor fused to mNeonGreen, which increases in fluorescence upon membrane depolarization. The construct includes the Kv2.1 proximal-restriction (PR) motif to confine expression to the cell body, enabling compartment-specific voltage imaging. The proximal-restriction (PR) motif derived from the C‑terminus of the Kv2.1 potassium channel doesn't exist in Paramecium caudatum. Its function when introduced into caudatum has not been verified. This specific pAce is codon optimized for P.caudatum replacing rare codon usage (<20%) with frequent codons from a newly computed P.caudatum codon table (43c3d annotation/assembly v1) with 64'003 coding sequences from 18'509 transcripts compromised of 8'507'122 codons. The pAce coding sequence is inserted between two flanking 5'UTR and 3UTR from the P.caudatum alpha-tubulin gene. The vector, pSF-aTubCaud, is a modification of the original phagemid pTZ18U vector. Before transfection by microinjection the plasmid is linearized using ApaI restriction enzyme. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 30°C Strain: NEB Stable |
TBD |
| pSF-aTaubCaud-FLAG-GCaMP8s |
TBD | GCaMP8s plasmid for generating calcium indicator-expressing transgenic Paramecium caudatum. GCaMP8s is a slow-speed, genetically encoded calcium indicator consisting of a circularly permuted EGFP (cpEGFP) fused to calmodulin (CaM) and an M13 peptide. Calcium binding triggers CaM–M13 interaction, inducing a conformational change that enhances cpEGFP fluorescence. GCaMP8s slow kinetics, high sensitivity, and an improved signal-to-noise ratio, making it well-suited for detecting slow calcium transients in neurons. Instead of a 6xHis-tag the construct has a FLAG-tag. This specific GCaMP8s codon optimized for P.caudatum replacing rare codon usage (<20%) with frequent codons from a newly computed P.caudatum codon table (43c3d annotation/assembly v1) with 64'003 coding sequences from 18'509 transcripts compromised of 8'507'122 codons. The GCaMP8s coding sequence is inserted between two flanking 5'UTR and 3UTR from the P.caudatum alpha-tubulin gene. The vector, pSF-aTubCaud, is a modification of the original phagemid pTZ18U vector. Before transfection by microinjection the plasmid is linearized using ApaI restriction enzyme. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 30°C Strain: NEB Stable 👩🏼💻 Examine plasmid directly in web-browser (SeqViz) 🧬 Download GeneBank file |
TBD |
| pSF-aTaubCaud-FLAG-GCaMP8m |
TBD | GCaMP8m plasmid for generating calcium indicator-expressing transgenic Paramecium caudatum. GCaMP8m is a medium-speed, genetically encoded calcium indicator consisting of a circularly permuted EGFP (cpEGFP) fused to calmodulin (CaM) and an M13 peptide. Calcium binding triggers CaM–M13 interaction, inducing a conformational change that enhances cpEGFP fluorescence. GCaMP8m offers balanced kinetics, high sensitivity, and an improved signal-to-noise ratio, making it well-suited for detecting moderate-speed calcium transients in neurons. Instead of a 6xHis-tag the construct has a FLAG-tag. This specific GCaMP8m codon optimized for P.caudatum replacing rare codon usage (<20%) with frequent codons from a newly computed P.caudatum codon table (43c3d annotation/assembly v1) with 64'003 coding sequences from 18'509 transcripts compromised of 8'507'122 codons. The GCaMP8m coding sequence is inserted between two flanking 5'UTR and 3UTR from the P.caudatum alpha-tubulin gene. The vector, pSF-aTubCaud, is a modification of the original phagemid pTZ18U vector. Before transfection by microinjection the plasmid is linearized using ApaI restriction enzyme. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 30°C Strain: NEB Stable |
TBD |
pSF-aTaubCaud-FLAG-GCaMP8f |
TBD | GCaMP8f plasmid for generating calcium indicator-expressing transgenic Paramecium caudatum. GCaMP8f is a fast, genetically encoded calcium indicator composed of a circularly permuted EGFP (cpEGFP) fused to calmodulin (CaM) and an M13 peptide. Upon calcium binding, CaM interacts with M13, inducing a conformational change that increases cpEGFP fluorescence. Engineered for rapid kinetics, high sensitivity, and improved signal-to-noise ratio, GCaMP8f is optimized for detecting fast calcium transients in neuronal activity. Instead of a 6xHis-tag the construct has a FLAG-tag. This specific GCaMP8f codon optimized for P.caudatum replacing rare codon usage (<20%) with frequent codons from a newly computed P.caudatum codon table (43c3d annotation/assembly v1) with 64'003 coding sequences from 18'509 transcripts compromised of 8'507'122 codons. The GCaMP8f coding sequence is inserted between two flanking 5'UTR and 3UTR from the P.caudatum alpha-tubulin gene. The vector, pSF-aTubCaud, is a modification of the original phagemid pTZ18U vector. Before transfection by microinjection the plasmid is linearized using ApaI restriction enzyme. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 30°C Strain: NEB Stable |
TBD |
| CAG-NLS-GFP | 104061 | GFP localized to nucleus trough N-term NLS from SV40. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 30°C Strain: NEB Stable 👩🏼💻 Examine plasmid directly in web-browser (SeqViz) 🧬 Download SnapGene file addgene-plasmid-104061-sequence-220228 |
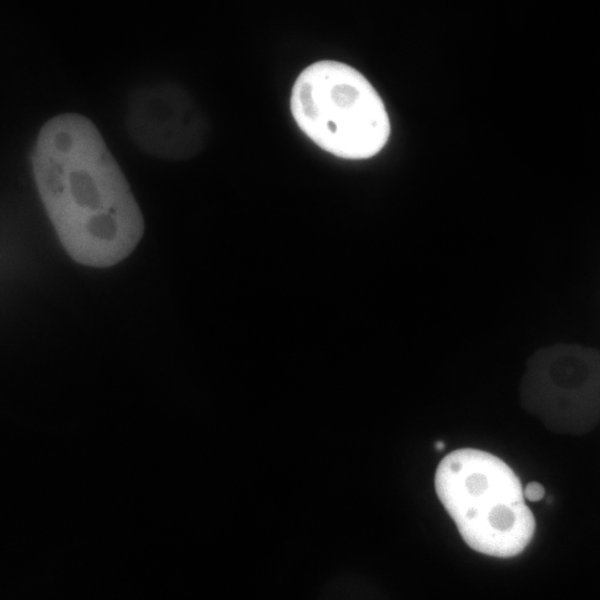 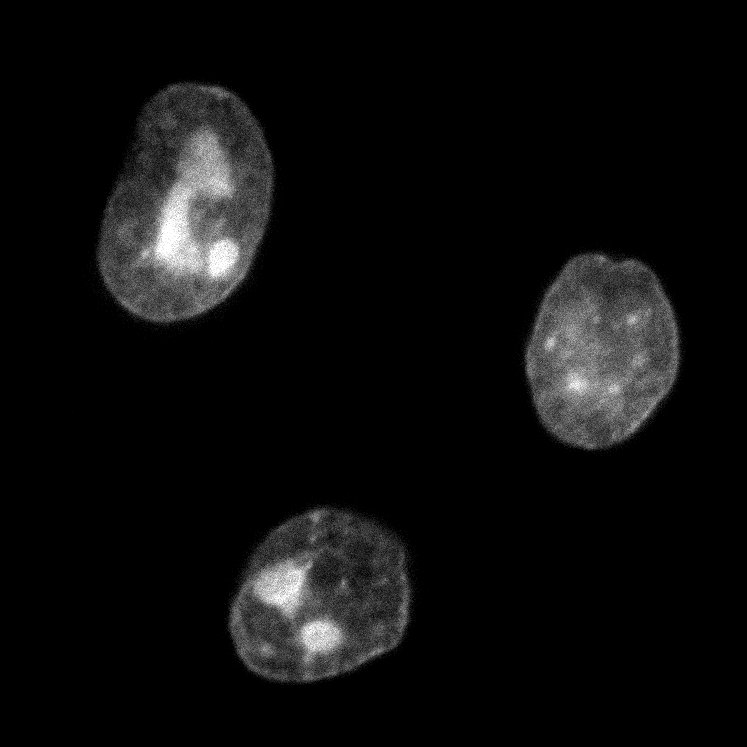
24h (left) and 72h (right). Notice the shift in signal to nucleoli. PC3 cells. |
| pCMV-GFP (pmaxGFP) | pmaxGFP | GFP, CMV promotor, a SV40 pA, a pUC ori and the kanamycin gene. |
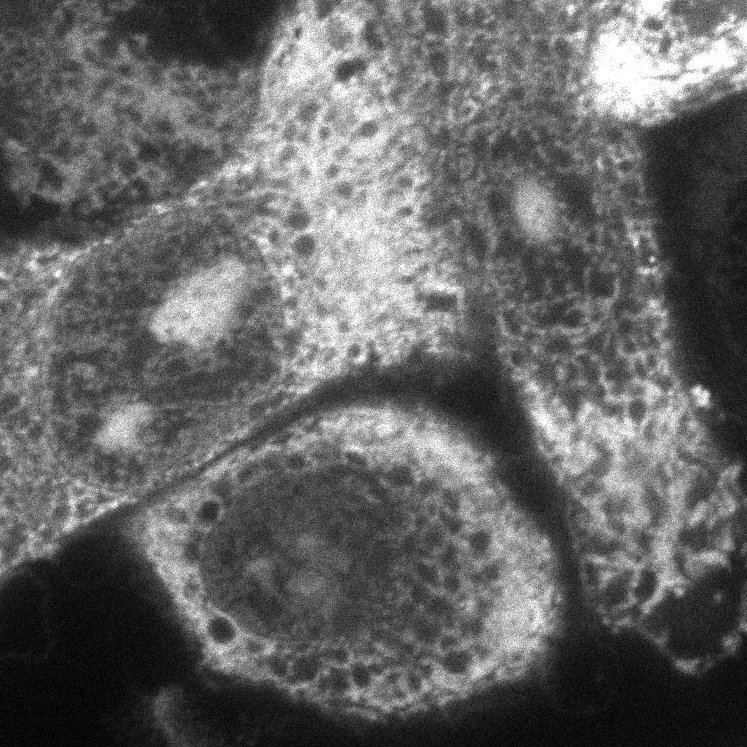
|
| pCAG-Palm-GFP | 14757 | GFP localized to membrane through N-term palmitoylation signal from GAP43. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 37°C Strain: DH5alpha 👩🏼💻 Examine plasmid directly in web-browser (SeqViz) 🧬 Download SnapGene file addgene-plasmid-28009-sequence-181212.dna |
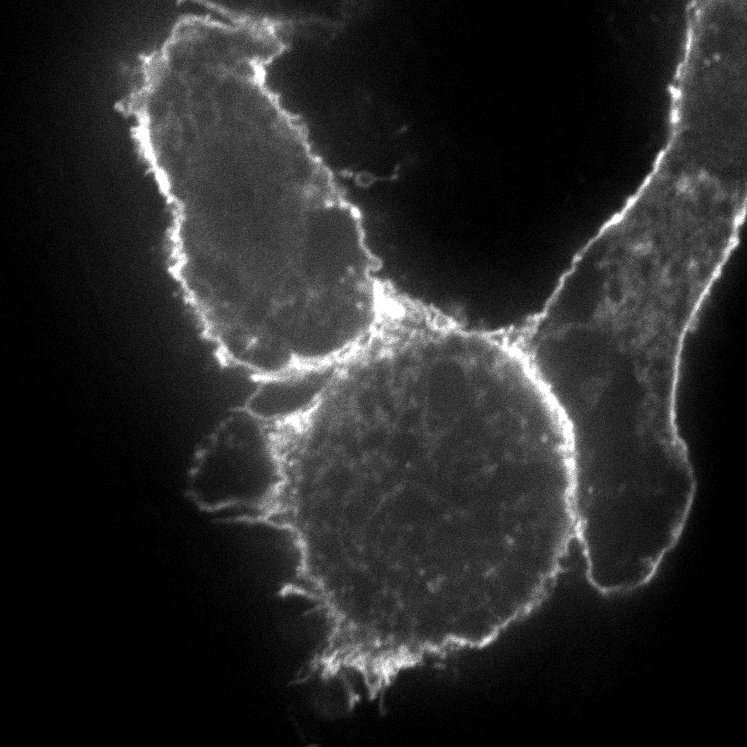
|
| CMV -Ecad-EGFP |
28009 | EGFP at C-terminal of Human Ecadherin in pcDNA3.1. The first two amino acids of the standard EGFP sequence (Met-Val) are missing due to the cloning strategy. There is also a Y238I mutation present at the C-terminus of EGFP. These differences do not affect the fluorescent protein properties. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 37°C Strain: DH5alpha 👩🏼💻 Examine plasmid directly in web-browser (SeqViz) 🧬 Download SnapGene file addgene-plasmid-28009-sequence-181212.dna |
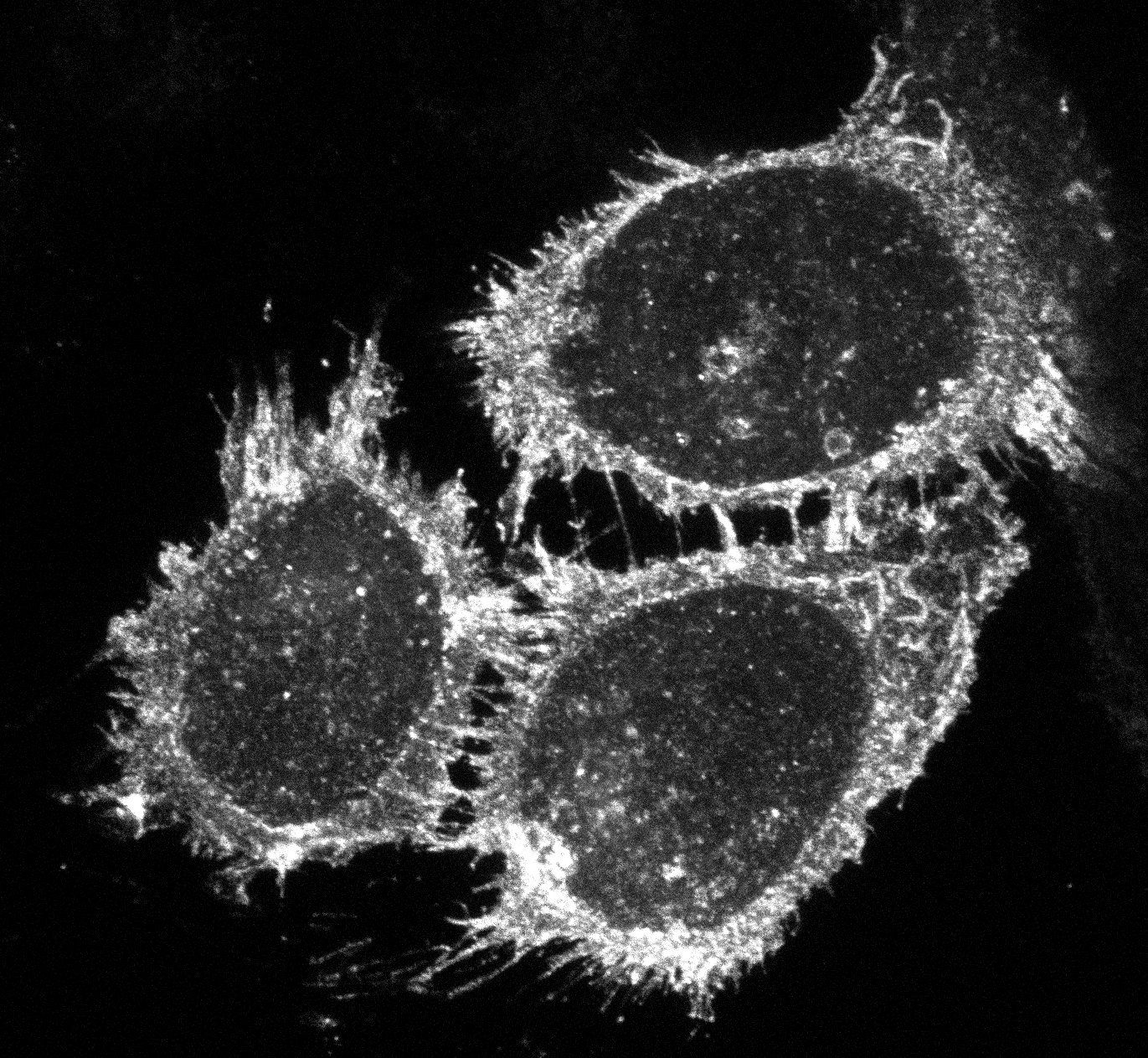
|
| pCMV-FLAG-Omomyc | 113168 |
Omomyc tagged with a FLAG-tag at the N terminal. Shown here stained with Rabbit anti-Flag as primary and Goat anti-Rabbit Alexa647 as secondary. Bacterial Resistance: Ampicillin, 100 μg/mL Growth Temperature: 37°C Strain: DH5alpha 👩🏼💻 Examine plasmid directly in web-browser (SeqViz) 🧬 Download SnapGene file addgene-plasmid-113168-sequence-217924.dna |
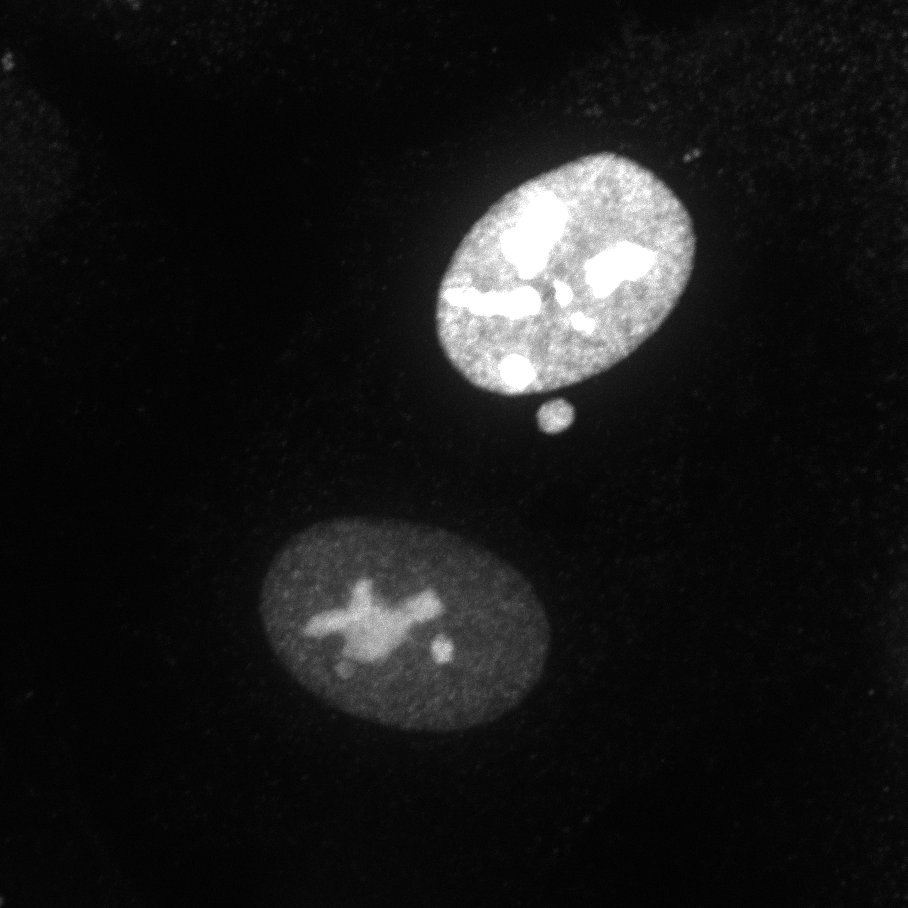
|
Universal sequencing primers
Universal primers play a crucial role in molecular biology techniques, particularly in Sanger sequencing and vector construction, by providing a reliable means of amplifying and analyzing a variety of sequences.
| Primer Name | Sequence (5' -> 3') | Length [bases] | Tm [°C] | GC [%] |
|---|---|---|---|---|
| -96gIII | CCC TCA TAG TTA GCG TAA CG | 20 | 57.3 | 50 |
| 1392r | ACG GGC GGT GTG TGT AC | 17 | 57.6 | 65 |
| 1492r | TAC GGT TAC CTT GTT ACG ACT T | 22 | 56.5 | 41 |
| 27f | AGA GTT TGA TCA TGG CTC A | 19 | 52.4 | 42 |
| 5AOX | GAC TGG TTC CAA TTG ACA AGC | 21 | 57.9 | 48 |
| ACYCDuetUP1 | GGA TCT CGA CGC TCT CCC T | 19 | 61.0 | 63 |
| BAC1 | GCT CGT ATG TTG TGT GGA ATT G | 22 | 58.4 | 45.5 |
| CMVfor | CGC AAA TGG GCG GTA GGC GTG | 21 | 65.7 | 67 |
| CMVmin | CGC CAT CCA CGC TGT TTT G | 19 | 58.8 | 58 |
| DuetDOWN1 | GAT TAT GCG GCC GTG TAC AA | 20 | 57.3 | 50 |
| DuetUP2 | TTG TAC ACG GCC GCA TAA TC | 20 | 57.3 | 50 |
| EGFP-Cfor | GAT CAC ATG GTC CTG CTG | 18 | 56 | 55.6 |
| EGFP-Nrev | CCG TCC AGC TCG ACC AG | 17 | 60 | 70.6 |
| EuVH | GGC AGC AGC CAC AGG TAA GA | 20 | 61.4 | 60 |
| EuVL | TTG CTG TTG CAC AGT GAT TC | 20 | 55.2 | 45 |
| Gal4AD | TAC CAC TAC AAT GGA TG | 17 | 47.9 | 41 |
| Gal4BD | TCA TCG GAA GAG AGT AG | 17 | 50.4 | 47 |
| HuCAL_VH_for | GAT AAG CAT GCG TAG GAG AAA | 21 | 55.9 | 43 |
| IgG_const_for | AGC CCA GCA ACA CCA AGG | 18 | 58.2 | 61 |
| IntXL39 | ATT AGG ACA AGG CTG GTG G | 19 | 56.7 | 53 |
| M13rev-29 | CAG GAA ACA GCT ATG ACC | 18 | 53.7 | 50 |
| M13rev-49 | GAG CGG ATA ACA ATT TCA CAC AGG | 24 | 61.0 | 46 |
| M13uni-21 | TGT AAA ACG ACG GCC AGT | 18 | 53.7 | 50 |
| M13uni-43 | AGG GTT TTC CCA GTC ACG ACG TT | 23 | 62.4 | 52 |
| pBABE3 | ACC CTA ACT GAC ACA CAT TCC | 21 | 57.9 | 48 |
| pBABE5 | CTT TAT CCA GCC CTC AC | 17 | 52.8 | 53 |
| pBAD-FP | ATG CCA TAG CAT TTT TAT CC | 20 | 51.1 | 35 |
| pBakPAC-FP | TAA AAT GAT AAC CAT CTC GC | 20 | 51.1 | 35 |
| pBR1 | CGA AAA GTG CCA CCT GAC | 18 | 56 | 55.6 |
| pBR3 | TCC CCA TCG GTG ATG TC | 17 | 55.2 | 58.8 |
| pcDNA3.1-RP_1 | CAA ACA ACA GAT GGC TGG C | 19 | 56.7 | 52.6 |
| pcDNA3_for | GGC TAA CTA GAG AAC CCA CTG | 21 | 59.8 | 52 |
| pcDNA3_rev | GGC AAC TAG AAG GCA CAG TC | 20 | 59.3 | 55 |
| pCEP-Forward | AGA GCT CGT TTA GTG AAC CG | 20 | 57.3 | 50 |
| pCEP-Reverse | GTG GTT TGT CCA AAC TCA TC | 20 | 55.2 | 45 |
| PCR2 | TTA GCT CAC TCA TTA GG | 17 | 47.9 | 41.2 |
| pCR3.1-BGHrev | TAG AAG GCA CAG TCG AGG | 18 | 56.0 | 56 |
| pDONOR-FP | TAA CGC TAG CAT GGA TCT C | 19 | 54.5 | 47.4 |
| pDONOR-RP | GCA ATG TAA CAT CAG AGA T | 19 | 50.2 | 36.8 |
| pEGFP_for | TTT AGT GAA CCG TCA GAT C | 19 | 52.4 | 42.1 |
| pEGFP_rev | TTT AAA GCA AGT AAA ACC TC | 20 | 49.1 | 30 |
| pENTattL1for | TCG CGT TAA CGC TAG CAT GGA TCT C | 25 | 64.6 | 52 |
| pENTattL2rev | ACA TCA GAG ATT TTG AGA CAC GGG C | 25 | 63.0 | 48 |
| pET-24a | GGG TTA TGC TAG TTA TTG CTC AG | 23 | 58.9 | 43.5 |
| petup | ATG CGT CCG GCG TAG A | 16 | 54.3 | 62 |
| pEX-For | GGA GCA GAC AAG CCC GTC AGG | 21 | 65.7 | 67 |
| pEX-Rev | CAG GCT TTA CAC TTT ATG CTT CCG GC | 26 | 64.8 | 50 |
| pFASTBAC-F | TCC GGA TTA TTC ATA CCG TCC C | 22 | 60.3 | 50 |
| pFASTBAC-R | CCT CTA CAA ATG TGG TAT GGC TG | 23 | 60.6 | 48 |
| pGexfor | ATA GCA TGG CCT TTG CAG G | 19 | 56.7 | 53 |
| pGEX-3 | GGA GCT GCA TGT GTC AGA GG | 19 | 58.8 | 57.9 |
| pGEX-5 | CTG GCA AGC CAC GTT TGG | 18 | 58.2 | 61.1 |
| pGEX5-FP | AAC GTA TTG AAG CTA TCC C | 19 | 52.4 | 42.1 |
| pGLrev | CTT TAT GTT TTT GGC GTC TTC C | 22 | 56.5 | 41 |
| pGL3for | CTA GCA AAA TAG GCT GTC CC | 20 | 57.3 | 50 |
| pIRES-RP | TAT AGA CAA ACG CAC ACC G | 19 | 54.5 | 47.4 |
| pJET1.2for | CGA CTC ACT ATA GGG AGA GCG GC | 23 | 66.0 | 61 |
| pJET1.2rev | AAG AAC ATC GAT TTT CCA TGG CAG | 25 | 66.0 | 52 |
| pJET1_fwd | GCC TGA ACA CCA TAT CCA TCC | 21 | 59.8 | 52 |
| pJet1-FP | ACT ACT CGA TGA GTT TTC GG | 20 | 55.3 | 45 |
| pJet1-RP | TGA GGT GGT TAG CAT AGT TC | 20 | 55.3 | 45 |
| pLKO1 | GAC TAT CAT ATG CTT ACC GT | 20 | 53.2 | 40 |
| pMalE | TCA GAC TGT CGA TGA AGC | 18 | 53.7 | 50 |
| Polyhed_fwd | AAA ATG ATA ACC ATC TCG | 18 | 46.9 | 33 |
| pQEfor | GTA TCA CGA GGC CCT TTC GTC T | 22 | 62.1 | 55 |
| pQErev | CAT TAC TGG ATC TAT CAA CAG GAG | 24 | 59.3 | 42 |
| pQE-FP | CGG ATA ACA ATT TCA CAC AG | 20 | 53.2 | 40 |
| pQE-RP | GTT CTG AGG TCA TTA CTG G | 19 | 54.5 | 47.4 |
| pRSET-RP | ATG CTA GTT ATT GCT CAG C | 19 | 52.4 | 42.1 |
| pTeSp-1 | CCT CCA TAG AAG ACA CC | 17 | 52.8 | 52.9 |
| pTrcHis-RP | CTG ATT TAA TCT GTA TCA GG | 20 | 51.1 | 35 |
| SP6 | CA TTT AGG TGA CAC TAT AG | 19 | 50.2 | 37 |
| T3 | AAT TAA CCC TCA CTA AAG GG | 20 | 53.2 | 40 |
| T7 | TAA TAC GAC TCA CTA TAG GG | 20 | 53.2 | 40 |
| T7term | CTA GTT ATT GCT CAG CGG T | 19 | 54.5 | 47 |
| T7-pET-mod | CCC GCG AAA TTA ATA CGA CTC AC | 23 | 60.6 | 48 |
| Topo-1 | TCG GAT CCA CTA GTA ACG | 18 | 53.7 | 50 |
| U6_fwd | GAG GGC CTA TTT CCC ATG ATT CC | 23 | 62.4 | 52.2 |
| v5epitoperev | CGT AGA ATC GAG ACC GAG GAG AGG | 24 | 66.1 | 58 |

